

Several experiments proved that DNA origami nanostructures possess abilities to enhance efficacies of chemotherapy, reduce adverse side effects, and even circumvent drug resistance. This breakthrough technique in structural DNA nanotechnology provides an easier and faster way to construct DNA nanostructures with various shapes. Such applications have become possible with the advent of the scaffolded DNA origami method. DNA, a biomolecule with molecular self-assembly property, emerges as a versatile nanomaterial to construct multi-functional platforms, which DNA nanostructures can be modified with functional groups to improve their utilities as biosensors or drug carriers. With nanocarriers, chemotherapeutic drugs could be directly delivered into target cancer cells resulting in enhanced efficiency with less side effects. Also, targeted drug delivery and precision medicine have now become a new paradigm in cancer therapy. Recently, cancer nanomedicine has gained much interest as promising diagnostic and therapeutic strategies as a wide range of nanomaterials possess unique physical properties that can render drug delivery systems more effective and safer.
#Capto butyl impres full#
The guide for creating advanced design motifs and the detailed protocols with their experimental characterization that we describe herein should lower the barrier for researchers to accomplish the full DNA origami production workflow.ĭue to the complexity and heterogeneity of cancer, the development of cancer diagnosis and therapy is still progressing, and a complete understanding of cancer biology still remain elusive.

These methods are agarose-gel purification, filtration via molecular cut-off membranes, PEG precipitation, size-exclusion chromatography, and ultracentrifugation-based sedimentation. In addition, we provide detailed protocols and discuss the expected results for five key methods that allow the efficient and damage-free preparation of DNA origami.
#Capto butyl impres how to#
We discuss design solutions for creating advanced structural motifs including corners and various types of hinges that expand the design space for the more rigid multi-layer DNA origami provide guidelines for preventing undesired aggregation and how to induce specific oligomerization of multiple DNA origami building blocks. To foster faster progress, in this article we share the experience that we have accumulated over the last years in making and preparing DNA origami. Such know-how is not readily available for newcomers to the field, thus slowing down the rate at which new applications outside the field of DNA nanotechnology may emerge. While the basic concept of DNA origami is easy to understand, using custom DNA origami in practical applications requires detailed know-how for designing and producing the particles with sufficient quality, and preparing them at appropriate concentrations with the necessary degree of purity in custom environments.

Gel electrophoresis showed high assembly yield and purity, whereas fluorescence correlation spectroscopy confirmed that the tetrahedrons had a diffusion coefficient (26.7 μm² s⁻¹) consistent with the expected size (20 nm).ĭNA ORIGAMI has attracted substantial attention since its invention ten years ago due to the seemingly infinite possibilities that it affords for creating customized nanoscale objects. In either case, collected ssDNA-containing fractions were homogeneous and impurity free.įinally, 8.4 μg of a 1000-nt ssDNA fragment was purified and used alongside with site-specific short oligonucleotides (staples) to assemble 63-bp edge length tetrahedrons.
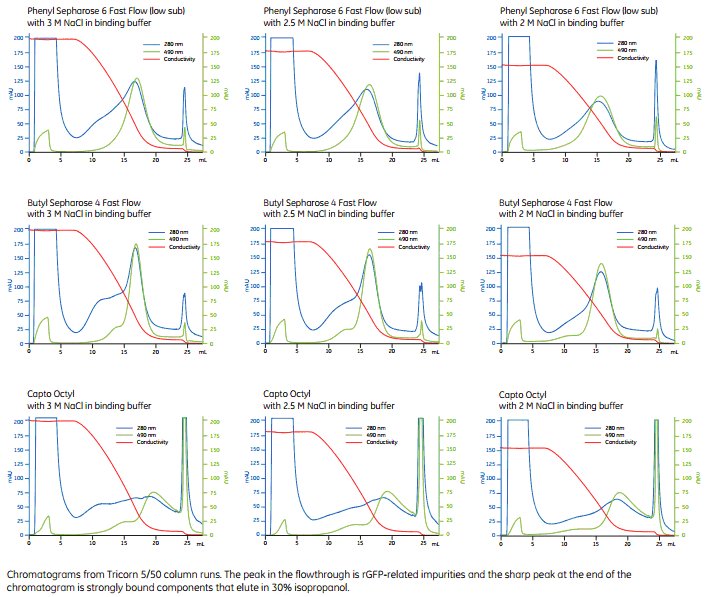
In multimodal chromatography, however, the elution pattern was reversed, highlighting the importance played by hydrophobicity. In anion exchange chromatography, the less-charged ssDNA eluted before the dsDNA. To isolate the target ssDNA from dsDNA and other PCR impurities, anion-exchange (Q-ligand) and multimodal chromatography (CaptoTM adhere ImpRes) were explored using stepwise gradients with increasing NaCl concentrations. Alternatively, we present a chromatography-based method to purify ssDNA scaffolds from aPCR mixtures, which can be used in the context of DNA-origami techniques.ĪPCR was performed to generate single and double-stranded DNA (dsDNA) from the M13mp18 genome. Each scaffold is usually purified by agarose gel extraction, a technique that is laborious, limited, not scalable, presents low recovery yields and a low-quality product. DNA-origami biomanufacturing relies in many cases on the use of asymmetric PCR (aPCR) to generate 500-3500 base, object-specific, single-stranded DNA (ssDNA) scaffolds.


 0 kommentar(er)
0 kommentar(er)
